rpart Decision tree for clustering in R
Before start of any analysis we need to check our dataset as it described here, in the section of the data Data Preparing for Cluster analysis
For using decision tree we need to use library rpart
> library(dplyr)
> library(rpart)Training and test model
When we have a lot of data, it is easier to select randomly test set by specifying the amount of percent of data in the and training sets. From my previous experience, i’ve find out, that 5% for test set is a good enough for many kind of statistical analysis
> iris_train <- iris %>% sample_frac(0.95)
> iris_test <- iris %>% setdiff(iris_train)As a result, it will be 142 observations in the train set and only 7 observations in the test set.
Decision tree
Calculate decision tree
We will calculate decision tree (rpart())of relation of Species to Petal length and Petal width of irises. And then we will draw the decision tree (plot()) and label everything (text)
> iris_tree2 <- rpart(Species ~ Petal.Length + Petal.Width,
+ data = iris_train,
+ method = "class")
> plot(iris_tree2, uniform = TRUE, margin = 0.5)
> text(iris_tree2, use.n = TRUE)After this code we will have this tree
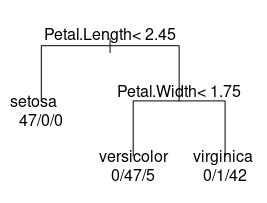
Clustering decision tree build with rpart()
As you can see, setosa was separated on the basis of Petal.Length and into this group all 47 points were fitted ideally. versicolor and virginica were separated on the basis of Petal.Width and this group is slightly mixed. versicolor has 47 correct and 5 wrong samples and virginica has 42 correct and 1 wrong data point
Predict with decision tree
We can use this decision tree to predict our test set. We will use our calculated decision tree iris_tree2 to apply it towards iris_test set with function predict() and we will calculate probability of each variant type = "prob" and also will ask about classification according to these probabilities vector type = "class"
> iris_test["Predict"] <- predict(iris_tree2, iris_test, type = "class")
> iris_test["Predict1"] <- predict(iris_tree2, iris_test, type = "prob")
> iris_test
Sepal.Length Sepal.Width Petal.Length Petal.Width Species Predict
1 5.1 3.7 1.5 0.4 setosa setosa
2 5.2 4.1 1.5 0.1 setosa setosa
3 5.0 3.5 1.6 0.6 setosa setosa
4 5.0 2.0 3.5 1.0 versicolor versicolor
5 6.3 2.3 4.4 1.3 versicolor versicolor
6 6.3 2.9 5.6 1.8 virginica virginica
7 7.7 2.6 6.9 2.3 virginica virginica
Predict1.setosa Predict1.versicolor Predict1.virginica
1 1.00000000 0.00000000 0.00000000
2 1.00000000 0.00000000 0.00000000
3 1.00000000 0.00000000 0.00000000
4 0.00000000 0.90384615 0.09615385
5 0.00000000 0.90384615 0.09615385
6 0.00000000 0.02325581 0.97674419
7 0.00000000 0.02325581 0.97674419As we can see all data are classified perfectly
Published: 2021-11-17 13:23:19
Updated: 2021-11-17 13:49:01