Data Preparing for Cluster analysis in R
Before clustering itself, we need to check dataset first. We need to check what this is (help()), how it is organised (glimpse()), any missed data (anyNA()) and also, if it is possible – visually check it, how it looks like (plot).
Dataset inspection
We will use internal, build it data set with iris species measurement iris
> help(iris)
iris package:datasets R Documentation
Edgar Anderson's Iris Data
Description:
This famous (Fisher's or Anderson's) iris data set gives the
measurements in centimeters of the variables sepal length and
width and petal length and width, respectively, for 50 flowers
from each of 3 species of iris. The species are _Iris setosa_,
_versicolor_, and _virginica_.
. . .
> glimpse(iris)
Rows: 150
Columns: 5
$ Sepal.Length 5.1, 4.9, 4.7, 4.6, 5.0, 5.4, 4.6, 5.0, 4.4, 4.9, 5.4,...
$ Sepal.Width 3.5, 3.0, 3.2, 3.1, 3.6, 3.9, 3.4, 3.4, 2.9, 3.1, 3.7,...
$ Petal.Length 1.4, 1.4, 1.3, 1.5, 1.4, 1.7, 1.4, 1.5, 1.4, 1.5, 1.5,...
$ Petal.Width 0.2, 0.2, 0.2, 0.2, 0.2, 0.4, 0.3, 0.2, 0.2, 0.1, 0.2,...
$ Species setosa, setosa, setosa, setosa, setosa, setosa, setosa...
> anyNA(iris)
[1] FALSE As we can see, this is a full dataset.
Plotting 2D datasets
We will use plot() to display our data
> plot(iris$Petal.Length,
+ iris$Petal.Width,
+ pch=c(20,22,24)[unclass(iris$Species)],
+ bg=c("red","green","blue")[unclass(iris$Species)])pch - give numbers if dot shapes and link them with variable Species, and bg - identify the colour of the selected dots.
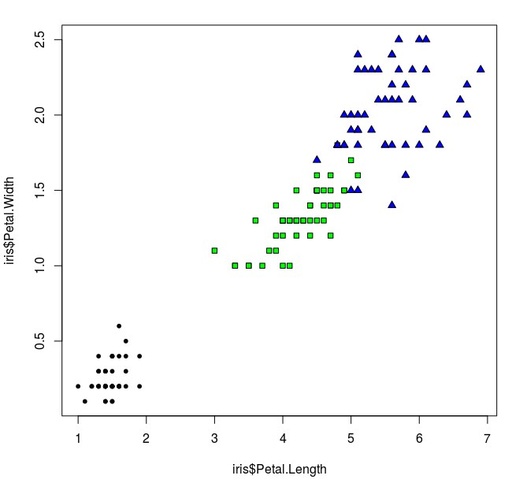
The dependence of Iris Petal.Length and Iris Petal.Width
Original image: 650 x 613
As we can see – data are grouped into 3 different clusters and now we need to calculate it statistically.
Preliminary analysis of the datasets
It is a good idea to manually check unlabelled dataset, to see is there are any groupping patterns is present
> unlabeled_iris <- iris %>% select(-Species)
> pairs(unlabeled_iris)This script will create table unlabeled_iris with removed species. So it will be anonymous dataset. And we will plot all possible pairs there.
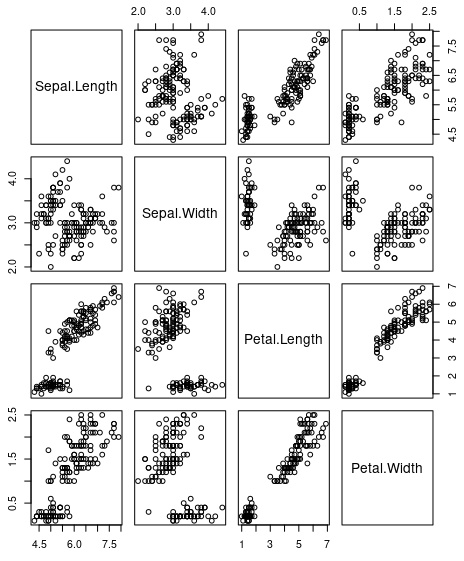
Anonymous pairs for iris dataset. Visual analysis should provide sme idea about grouping this data
Visual inspection of this pairs should give some ideas about possible clustering data for automatic learning.
Published: 2021-11-17 08:22:34
Updated: 2021-11-17 14:04:37